Data Transformation Application
This project involved developing software to transform data for a clinical application. The transformation that this software performs is for patient data in long form into wide form. If you want to skip to the section which details the clinical applications in which this software has been used, see the deployment section.
The following sections explain what the software does and the main problem the software solves. An explanation of the transformation process and the algorithm developed is also detailed. The final section provides the source code for this software.
What does the software do?
The software application performs the transformation of data from long to wide form. The basic idea behind the transformation process is to take all entries for a patient in long format, which are expressed as multiple rows, and produce a single row for a patient. Each of the values which are in rows in long form, will now be expressed as columns in wide format. The images below show an example transformation from long form to wide form data.
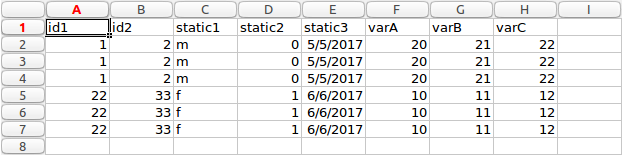
Long form
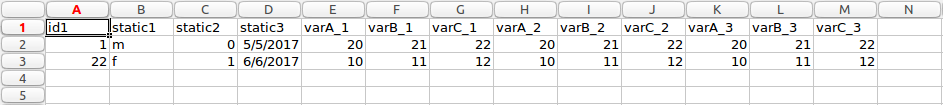
Wide form
It can be seen from the images above the transformation process is not a simple as each row in long form is expressed as a column in wide form. There are static and variable columns, as well as multiple unique ids and columns to ignore. For this reason, the particular application discussed here takes ‘complex’ long form data and convert to wide form.
What do we mean by complex long form data?
The process of converting from long to wide is not simply the case of taking all entries expressed as a row, and outputting them as columns. We have to consider that some values do not need to be express as multiple columns, and also we have some values which need to be ignored.
The Problem
In order to explain the problem, it is first necessary to explain the data we are dealing with in this particular application. The data is motivated by a problem in the clinical environment, which influences the terminology of the explanation.
Given the example data in long form, it can be see we have two different patients, highlighted as orange and blue.
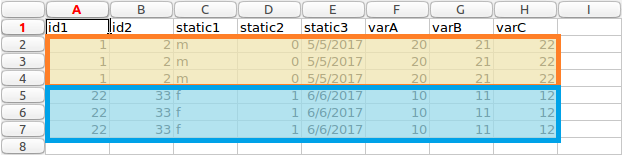
Patients highlighted long form data
In this data, each patient has a number of associated values. They have two identifiers, id1 and id2 in columns A and B. These values are unique to a patient. They also have some other values. We define these in two categories, either static or variable. The static values stay the same for each row for each patient. You can see this by looking at columns C, D and E. If you look at rows 2,3 and 4, which are for the first patient (highlighted as orange in image above), we can see the values stay the same for each entry.
The variable values can change for each row. In this example, they would be columns F,G and H. They are not different in this example however, this is for ease of explanation and testing if values are mapped correctly from wide to long format.
The resulting transformed file is shown below. The first patient is highlighted in orange, and the second in blue, as above. This particular example was transformed using the following parameters, more information about the type of parameters this software uses can be read in transformation process section.
idColumn=0 staticColumns=[2,3,4] ignoreColumns=[1]
It can be seen how the data format has changed from long to wide format.
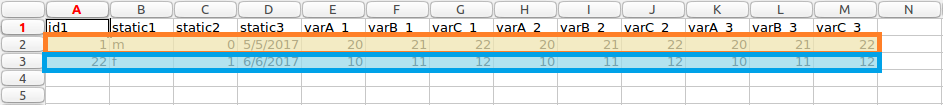
Patients highlighted wide form data
In order to give some context to this data, I am going to give an example of the data that could be in this format. Every time a patient has some clinical values recorded, a new row is added to a data set. There are two ids because two systems are used to record different data about a patient. The static values are value which typically do not change. This could be values like date of birth and gender. This could also be certain disease categories. The variable values could then relate to the clinical measurements taken at a point in time. This could be a blood glucose measurement, weight, or a certain treatment for example.
The transformation process needs to take all of these individual entries express as rows, and produce a single row in the wide form data output.
Transformation Process
The transformation process from long form to wide form has some important concepts that need to be understood before the algorithm can be explained. The concepts are:
- Id column
- Static columns
- Ignore columns
Id column This is needed in order to uniquely identify each of the individual patients rows in the long form data.
Static column Static columns are identified such that they do not contain different values in the long form of the data. This means that we only need to have each of these values expressed as one column in the single entry wide form data.
In the example shown, columns C,D and E contain the exact same values for a patient. As a result, they do not need to be expressed as multiple columns in wide form.
Ignore column Ignore columns are any columns that are not required in the wide form data output. Using our example, this is the extra id2 column. But could be any value.
Algorithm
The transformation algorithm has two important parts in its main loop. Each of these two parts will be explained with code samples below. The main loop iterates for each row in the long form input data file. The function of the main loop is to build up a single row of data for each of the multiple rows of data read from the long form file. Once the single row of data is constructed for a patient, it is output to file, and the row of data for the next patient is constructed, and so on. As mentioned, there are two important parts for this process to function.
Part 1
The first part within the main loop is used to determine if the current row relates to a new patient or the previous patient. This translates to if the current row is the same patient as the previous row of data or not.
if not then;
- write the current row data to file it is not empty
- reset the current row data
- update maxIterations by comparing the currentIterations. This is to know what the max number of possible columns are going to be in the wide format. Which is essentially determined by the max number of rows for all patients
- Append the current values for the start of the next persons wide form data. This includes adding the id value, and any static values
- move onto part 2
if yes then;
- increment the current iteration
- move onto part 2
The code below shows how this part works.
idValue = row[idColumn]
print("idvalue="+idValue)
if idValue != currentId:
print(" new person")
# write current row data (if not empty)
if rowData != []:
print(" Write previous persons data "+str(currentIteration))
print(rowData)
write.writerow(rowData)
# reset row data
rowData=[]
# update max iterations if more
if currentIteration > maxIteration:
maxIteration=currentIteration
# reset iteration count
currentIteration=1
# set current id value
currentId=idValue
# write new row details
# (1) write id
rowData.append(idValue)
# (2) write static columns
for staticItem in staticColumns:
#print(staticItem)
rowData.append(row[staticItem])
else:
print(" not new person")
currentIteration=currentIteration+1
Part 2
The second part within the main function acts to check if there are any columns that need to be excluded. This is because they are contained in the set of ignore columns. Or it could be because it is static data, and does not need to be repeated. Remember, the static data for the patient has already been output at the start of the single row of data in part 1 (as described above).
if the column is not in the static or ignore sets, then it is to be append to the current row data. The main loop then continues.
The code below shows how this part works.
for i in range(0,len(row)):
# check if we should ignore this row
if i == idColumn:
print(" dont output "+str(i))
elif i in staticColumns:
print(" dont output "+str(i))
elif i in ignoreColumns:
print(" dont output "+str(i))
else:
# append item to current row
print(" printing "+str(i))
rowData.append(row[i])
Main loop
The full code for the main loop is shown below. This includes parts 1 and 2, as explained above, and also the initial setup of counters and the processes that are done at the end of the final loop.
currentIteration=1
currentId=0
rowData=[]
maxIteration=0
#For each row
for row in read:
#For each column in row
idValue = row[idColumn]
print("idvalue="+idValue)
# Part 1
if idValue != currentId:
print(" new person")
# write current row data (if not empty)
if rowData != []:
print(" Write previous persons data "+str(currentIteration))
print(rowData)
write.writerow(rowData)
# reset row data
rowData=[]
# update max iterations if more
if currentIteration > maxIteration:
maxIteration=currentIteration
# reset iteration count
currentIteration=1
# set current id value
currentId=idValue
# write new row details
# (1) write id
rowData.append(idValue)
# (2) write static columns
for staticItem in staticColumns:
#print(staticItem)
rowData.append(row[staticItem])
else:
print(" not new person")
currentIteration=currentIteration+1
# Part 2
for i in range(0,len(row)):
# check if we should ignore this row
#
if i == idColumn:
print(" dont output "+str(i))
elif i in staticColumns:
print(" dont output "+str(i))
elif i in ignoreColumns:
print(" dont output "+str(i))
else:
# append item to current row
print(" printing "+str(i))
rowData.append(row[i])
# write last persons row data (if not empty)
if rowData != []:
print(" Write LAST persons data "+str(currentIteration))
print(rowData)
write.writerow(rowData)
Deployment
This section describes two examples in which this application has been deployed in real world scenarios in the healthcare sector.
- Diabetic Eye Screening Analysis
- Blood Glucose Analysis
Diabetic Eye Screening
The first example of this application being deployed is for the analysis of diabetic eye screening and GP data. The long form data is such that each row contains recorded clinical values from an eye screening examination along with the nearest clinical measurements recorded at a GP.
The software application was used to transform the long form data, such that all records for a single patient expressed as rows, were then processed as expressed in multiple columns, as a single row for each patient.
Blood Glucose
The second example of this application being deployed was for detailed analysis of a certain single covariate from a wealth of GP data. The value wanted for analysis in this particular case was HbA1c which is a measurement of blood glucose level. The various HbA1c value were expressed as a new row for each measurement taken. This data also contained records for some 30+ other covariates, along with some static values.
The software application was used to transform the long form data described above into wide form data, such that only HbA1c values were in multiple columns, as a single row for each patient.
Source Code
Update 2018-01-16. A features was added so that duplicates can be removed from the input file. For source code see github.com/aalshukri/PythonDataTransformApp
Usage
The code below shows an example usage of the software. This will transform the file testdata1.csv into the form testdata1_wide.csv
# filename
longFormFileName = 'testdata/testdata1.csv'
wideFormFileName = 'testdata/testdata1_wide.csv'
# id column
idColumn=0
# staticColumns
staticColumns=[2,3,4]
# ignoreColumns
ignoreColumns=[1]
The starting long form and resulting wide form is shown below.