Segmentation and batch processing with legacy data systems
In the healthcare environment old data system are often relied upon as they are heavily integrated in patient pathways. When these systems are to integrate with newer systems, they are often referred to as legacy systems. The legacy systems were designed for a specific purpose and scope. With the ever growing data generation and complexity, these systems are not fit for the sudden increase in data demands. Despite this increase in demand, they are crucial and heavily integrated into the current systems and processes. There systems are still need to be utilised as part of a clinical trials and for other supporting research (as in this case).
In the field of data engineering we are often faced with such problems. In this work, in order to deal with the problem of using legacy system with much larger data demands, an application was developed to interface with legacy data processing systems. This application was used for segmentation and batch processing with legacy data systems.
The specific problem which this application solved is explained further.
Problem
The problem was that we heavily relied on legacy code bases for data merging and transformation. Theses system were designed and worked well for amounts of data at the time of development. But they do not scale well to the larger amounts of data we have today. There was no short term alternative to the legacy code bases. In order to interface with the legacy systems and deal with the much larger data demands an application was developed for segmentation and batch processing.
Legacy system: data merging
In order to explain the segmentation solution, it is first necessary to understand the function of the legacy system. For this explanation, the merge process from a legacy system with be the focus.
This will be explained using generic terminology. We have a ‘main file’, and some number of ‘supporting files’ (sup files). The output of the legacy system here is such that; each record of the main file, merged with information found somewhere in the supporting files.
Originally, when running this process using the legacy systems, the full data set could be imported and loaded into memory. The system could then be used to run the various data merges to calculate the final output. Using the full data sets was no problem at the time.
When data is large, there is no way of doing this, hence we need to segment the input files and batch process.
Solution
This section describes the solution developed to solve the problem outlined above. The focus will be on how the segmentation process works, and the two types of segmentation that can be performed as part of the application developed in this work.
Segmentation Process
There are two main methods of segmentation and batch processing used in this work.
- Take the main file, and segment supporting files. Known as Single Segmentation.
- Segment the main file, and then produce segmented supporting file for each segmented base file. Know as Multi Segmentation.
Single Segmentation
In the single segmentation process, the main file is used to segment the supporting files.
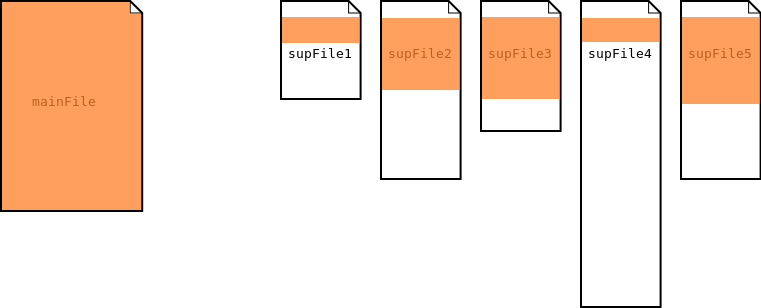
Single Segmentation
Multi Segmentation
In the multi segmentation process, the main file is first segmented. Then each of these segments is used to further segment each of the supporting files
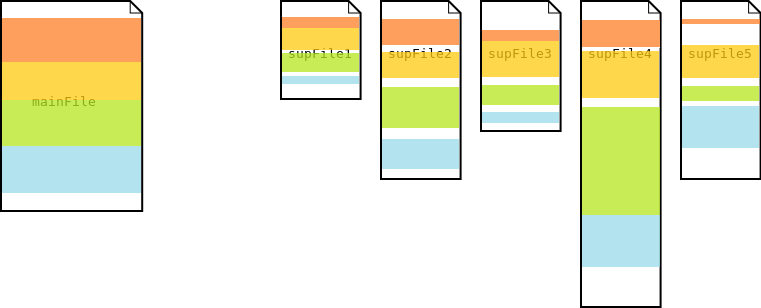
Multi Segmentation
Assumptions
The segmentation process explained above assumes that the files are in sorted order by id. This is actually a naive approach to segmentation. The reason is that if we cannot guarantee the main file is sorted by ids, then we could in fact end up with a segmentation which is exactly the same as the set of input files in terms of size and complexity. This would be bad for two reasons:
- The processing for segmentation to output files would be a waste
- The legacy code would simply not work with the segmented files even when used in a batch processing scenario
As an example. consider that there are 4 unique ids. If these ids are repeated in the main file such that they are distributed in an uneven way. We could segment our main file, such that each segment contains one of the 4 ids. This will mean the subsequent segmentation process will not produce files which have a reduction in file size. This means the output would still contain the complexities of the input.
Software
This section outlines some interesting and novel aspects of the software application. The particular aspects of interest in this application were the ‘Chunking Algorithm’.
Chunking Algorithm
This function reads an input file and chunks it into separate files based on the numChunksVariable and the number of rows in the input file.
Algorithm
This algorithm iterates over each line in the input file and outputs the line to a new output file based on the:
- number of chunked output files specified
- while preserving a complete set of ids is not split across multiple files
The algorithm works by first calculating the number of rows in the input file. This is done by iterating over all lines in the file. This is scalable to large files as the file data is not held in memory. Using this number, and the given numChunks variable, the number of items per chunk can be calculated.
numRowsPerChunk = totalNumRows / numChunks
The input file is then iterated line by line outputting the rows to a new file. The remained of a division calculation is used to determine when a new segment should be created, and thus if a new file to output to should be created or not.
rowCounter % numRowsPerChunk
The remained of the calculation is 0 if the rowCounter is divisible exactly by the numRowsPerChunk. Which indicates that the rowCounter is at the end of the number of rows that should be added to a file.
Example of reminder calculation
A simple example of the remainder calculation is show below. The file has 6 rows, the number of chunks specified is 3. Therefore we should chunk the input file into 3 files, each containing 2 rows. Not considering headers.
numChunks = 3
totalNumRows = 6
numRowsPerChunk = 2
0
rowCounter(0) % numRowsPerChunk(2) = 0
NewFile 1
Write row to file num 1
1
rowCounter(1) % numRowsPerChunk(2) = 1
chunkFileCounter = 1
Write row to file num 1
2
rowCounter(2) % numRowsPerChunk(2) = 0
NewFile 2
Write row to file num 2
3
rowCounter(3) % numRowsPerChunk(2) = 1
Write row to file num 2
4
rowCounter(4) % numRowsPerChunk(2) = 0
NewFile 3
Write row to file num 3
5
rowCounter(5) % numRowsPerChunk(2) = 1
Write row to file num 3.csv
Uneven file distribution
The algorithm has a logical check to make sure that an uneven distribution of rows are still divided into the specified number of files given. The if statement ensures that any extra rows are simply added to the last file. This statement only created a new file while the number of files do not exceed the number of chunks.
if not chunkFileCounter>numChunks
Preserve ids in single files
The algorithm also have a logical check to make sure that a set of ids is not split across multiple files. This part of the algorithm helps preserve that a set of ids for a single patient is kept to one file, and is not partially split amongst different files.
This works by checking the value of the id column of the previous row. If the same and current row, then don’t split to a new file. If the current row value is different, then it is fine to split into different files. This feature is auto enabled for all file chunking using this methods as it is an important requirement for this application.
Pseudocode
The algorithm Pseudocode code which combines all of the concepts explained above in to the algorithm.
For each row in input
currentIdValue=row[columnId]
if rowCounter % numRowsPerChunk == 0:
chunkFileCounter=chunkFileCounter+1
if not chunkFileCounter>numChunks:
createNewFileFlag=True
if createNewFileFlag:
# if current and previous id not equal, then is a new person.
# and we not exceeding the max number of file chunks
# then create a new file
if (not previousIdValue == currentIdValue) and (not chunkFileCounter>numChunks):
currentFileName = update file name using chunkFileCounter
currentFileWriter = getNewFileWriter(currentFileName)
createNewFileFlag=False
currentFileWriter.writerow(row)
rowCounter=rowCounter+1
previousIdValue=currentIdValue
Endfor
Function output
This function returns list of chunkedFileNames as the output. This is then used for further segmentation processes in the main method of the application.
Examples
Some examples of the algorithm above in action on some sample files.
Example A
All data should be written to one single file. ie, no file chunks created.
main
chunkFile[data/testdata1/main.csv]
numChunks = 1
totalNumRows = 6
numRowsPerChunk = 6
0
rowCounter(0) / numRowsPerChunk(6) = 0
rowCounter(0) % numRowsPerChunk(6) = 0
chunkFileCounter = 0
NewFile
chunkFileCounter = 1
Write row to file num = seg1.csv
['001', 'aa', 'bb']
1
rowCounter(1) / numRowsPerChunk(6) = 0
rowCounter(1) % numRowsPerChunk(6) = 1
chunkFileCounter = 1
Write row to file num = seg1.csv
['001', 'aa', 'bb']
2
rowCounter(2) / numRowsPerChunk(6) = 0
rowCounter(2) % numRowsPerChunk(6) = 2
chunkFileCounter = 1
Write row to file num = seg1.csv
['003', 'aa', 'bb']
3
rowCounter(3) / numRowsPerChunk(6) = 0
rowCounter(3) % numRowsPerChunk(6) = 3
chunkFileCounter = 1
Write row to file num = seg1.csv
['003', 'aa', 'bb']
4
rowCounter(4) / numRowsPerChunk(6) = 0
rowCounter(4) % numRowsPerChunk(6) = 4
chunkFileCounter = 1
Write row to file num = seg1.csv
['004', 'aa', 'bb']
5
rowCounter(5) / numRowsPerChunk(6) = 0
rowCounter(5) % numRowsPerChunk(6) = 5
chunkFileCounter = 1
Write row to file num = seg1.csv
['004', 'aa', 'bb']
Example B
Data should be written to 2 files. Number of 6 lines should be chunked accross two files, seg1 and seg2
main
chunkFile[data/testdata1/main.csv]
numChunks = 2
totalNumRows = 6
numRowsPerChunk = 3
0
rowCounter(0) / numRowsPerChunk(3) = 0
rowCounter(0) % numRowsPerChunk(3) = 0
chunkFileCounter = 0
NewFile
chunkFileCounter = 1
Write row to file num = seg1.csv
['001', 'aa', 'bb']
1
rowCounter(1) / numRowsPerChunk(3) = 0
rowCounter(1) % numRowsPerChunk(3) = 1
chunkFileCounter = 1
Write row to file num = seg1.csv
['001', 'aa', 'bb']
2
rowCounter(2) / numRowsPerChunk(3) = 0
rowCounter(2) % numRowsPerChunk(3) = 2
chunkFileCounter = 1
Write row to file num = seg1.csv
['003', 'aa', 'bb']
3
rowCounter(3) / numRowsPerChunk(3) = 1
rowCounter(3) % numRowsPerChunk(3) = 0
chunkFileCounter = 1
NewFile
chunkFileCounter = 2
Write row to file num = seg2.csv
['003', 'aa', 'bb']
4
rowCounter(4) / numRowsPerChunk(3) = 1
rowCounter(4) % numRowsPerChunk(3) = 1
chunkFileCounter = 2
Write row to file num = seg2.csv
['004', 'aa', 'bb']
5
rowCounter(5) / numRowsPerChunk(3) = 1
rowCounter(5) % numRowsPerChunk(3) = 2
chunkFileCounter = 2
Write row to file num = seg2.csv
['004', 'aa', 'bb']
Example C
The number of input lines is 6, the number of chunks specified is 3. This means we should have 3 files, each with Number of 6 lines should be chunked accross three files, seg1, seg2 and seg3.
main
chunkFile[data/testdata1/main.csv]
numChunks = 3
totalNumRows = 6
numRowsPerChunk = 2
TEST numRowsPerChunk = 2.0
TEST numRowsPerChunk = 2.0
TEST numRowsPerChunk = 2
TEST numRowsPerChunk = (0.0, 2.0)
TEST numRowsPerChunk = 0.0
TEST numRowsPerChunk = 0.0
TEST numRowsPerChunk = 0
TEST numRowsPerChunk = (0.0, 0.0)
0
rowCounter(0) / numRowsPerChunk(2) = 0
rowCounter(0) % numRowsPerChunk(2) = 0
chunkFileCounter = 0
chunkFileCounter = 1
NewFile
data/testdata1/main.seg1.csv
Write row to file num = data/testdata1/main.seg1.csv
['001', 'aa', 'bb']
1
rowCounter(1) / numRowsPerChunk(2) = 0
rowCounter(1) % numRowsPerChunk(2) = 1
chunkFileCounter = 1
Write row to file num = data/testdata1/main.seg1.csv
['001', 'aa', 'bb']
2
rowCounter(2) / numRowsPerChunk(2) = 1
rowCounter(2) % numRowsPerChunk(2) = 0
chunkFileCounter = 1
chunkFileCounter = 2
NewFile
data/testdata1/main.seg2.csv
Write row to file num = data/testdata1/main.seg2.csv
['003', 'aa', 'bb']
3
rowCounter(3) / numRowsPerChunk(2) = 1
rowCounter(3) % numRowsPerChunk(2) = 1
chunkFileCounter = 2
Write row to file num = data/testdata1/main.seg2.csv
['003', 'aa', 'bb']
4
rowCounter(4) / numRowsPerChunk(2) = 2
rowCounter(4) % numRowsPerChunk(2) = 0
chunkFileCounter = 2
chunkFileCounter = 3
NewFile
data/testdata1/main.seg3.csv
Write row to file num = data/testdata1/main.seg3.csv
['004', 'aa', 'bb']
5
rowCounter(5) / numRowsPerChunk(2) = 2
rowCounter(5) % numRowsPerChunk(2) = 1
chunkFileCounter = 3
Write row to file num = data/testdata1/main.seg3.csv
['004', 'aa', 'bb']
Example D
The number of input lines is 7, the number of chunks specified is 3. This means we should have 3 files, each with 2 items, and the last remaining item will be places in the last file. which will contain 3 items. Number of 7 lines should be chunked accross three files, seg1, seg2 and seg3.
main
chunkFile[data/testdata1/main.csv]
numChunks = 3
totalNumRows = 7
numRowsPerChunk = 2 ~ totalNumRows not exactly divisable by numChunks. remainders added to last file.
TEST numRowsPerChunk = 2.0
TEST numRowsPerChunk = 2.0
TEST numRowsPerChunk = 2
TEST numRowsPerChunk = (0.0, 2.0)
TEST numRowsPerChunk = 1.0
TEST numRowsPerChunk = 1.0
TEST numRowsPerChunk = 1
TEST numRowsPerChunk = (0.0, 1.0)
0
rowCounter(0) / numRowsPerChunk(2) = 0
rowCounter(0) % numRowsPerChunk(2) = 0
chunkFileCounter = 0
chunkFileCounter = 1
NewFile
data/testdata1/main.seg1.csv
Write row to file num = data/testdata1/main.seg1.csv
['001', 'aa', 'bb']
1
rowCounter(1) / numRowsPerChunk(2) = 0
rowCounter(1) % numRowsPerChunk(2) = 1
chunkFileCounter = 1
Write row to file num = data/testdata1/main.seg1.csv
['001', 'aa', 'bb']
2
rowCounter(2) / numRowsPerChunk(2) = 1
rowCounter(2) % numRowsPerChunk(2) = 0
chunkFileCounter = 1
chunkFileCounter = 2
NewFile
data/testdata1/main.seg2.csv
Write row to file num = data/testdata1/main.seg2.csv
['003', 'aa', 'bb']
3
rowCounter(3) / numRowsPerChunk(2) = 1
rowCounter(3) % numRowsPerChunk(2) = 1
chunkFileCounter = 2
Write row to file num = data/testdata1/main.seg2.csv
['003', 'aa', 'bb']
4
rowCounter(4) / numRowsPerChunk(2) = 2
rowCounter(4) % numRowsPerChunk(2) = 0
chunkFileCounter = 2
chunkFileCounter = 3
NewFile
data/testdata1/main.seg3.csv
Write row to file num = data/testdata1/main.seg3.csv
['004', 'aa', 'bb']
5
rowCounter(5) / numRowsPerChunk(2) = 2
rowCounter(5) % numRowsPerChunk(2) = 1
chunkFileCounter = 3
Write row to file num = data/testdata1/main.seg3.csv
['004', 'aa', 'bb']
6
rowCounter(6) / numRowsPerChunk(2) = 3
rowCounter(6) % numRowsPerChunk(2) = 0
chunkFileCounter = 3
chunkFileCounter = 4
Write row to file num = data/testdata1/main.seg3.csv
['005', 'aa', 'bb']
Usage
There is some test data and a test configuration file included in the repository for this software.
The files contained:
- main.csv
- supA.csv
- supB.csv
- supC.csv
The config file:
[mainFile]
dir=data/testdata1
mainFileName = %(dir)s/main.csv
mainFileColumnId = 0
numChunks = 3
[supportingFiles]
dir=data/testdata1
supportingFileNames = %(dir)s/supA.csv
%(dir)s/supB.csv
%(dir)s/supC.csv
supportingFileColumnIds = 0
0
0
It is possible to use this program in two modes. These modes are switched using the ‘numChunks’ param in the mainFile config section. If no param is supplied, then there will be no chunking of the main input file. If a value is supplied, then this number will be used to split the main input file into equal segments.
Example command used to execute the program
> python FileSegmentationApp.py testdata1/config.ini
Code sample available FileSegmentationForBatchJobs.
Program Time and Memory Complexity Analysis
http://bigocheatsheet.com/
Time Complexity:
The time complexity of the algorithm is ~2n. n for reading the input and determining the size of the file. and n for iterating over the file in order to create the segments.
Possible solution to reduce complexity:
- Use a threshold value for the input file size. This could be an estimate based on input file size in bytes which can be access without reading lines of the file.
- How to iterative over input file less than
ntimes? Possibly use a divide and conqueror type approach. Start a middle of file, then determine segments? Not a solution, just an idea.
Memory The memory footprint is low. But I/O and processing is high. This decision was made in order to scale to large input file size. ie.
- The input files are never stored into memory.
- The output data is flushed periodically and not stored in memory. This requires more processing and I/O, but is saleable to large files sizes.